Our research group aims to understand key aspects of host-pathogen interactions. We explore a range of scales from molecular interactions, through pheno- and genotypes, to population levels, applying a systems biological approach. To do so we use techniques including classical epidemiology and immunology, cutting edge molecular techniques and high throughput pathogen genome sequencing. We work closely with hospitals and clinics around Switzerland, Europe and globally, giving us access to clinical samples and patient data, where required. We want to translate our findings into clinical practice, developing novel diagnostic strategies and preventive measurements to reduce pathogen transmission.
The Applied Microbiology Research Lab is located at the Institute of Medical Microbiology, University of Zurich
Research
Research Focus
Transmission of clinically relevant pathogens
Infectious diseases cause significant morbidity and mortality. Understanding the sources, transmission and dynamics of pathogens is key to identifying and preventing outbreaks.
Bacterial transmission models
Bacterial pathogens of interest include multi-drug resistant bacteria such as ESBL- and Carbapenemase-producing and hypervirulent Enterobacteriaceae, Clostridioides difficile, and Methicillin resistant Staphylococcus aureus, as well as interesting clinical outbreaks. We use whole genome sequencing (WGS; Illumina and Oxford Nanopore) and metagenomic approaches in order to describe genetic relatedness and evolution within hosts.
Specific projects include:
- Developing metagenomic tools to determine MDR colonization status of patients directly from swabs. Bioinformatic tools will be developed to monitor microbiota changes over time within the patient during hospitalisation.
- Investigating the transmission dynamics of ESBL plasmids in Enterobacteriaceae: in collaboration with colleagues from ETH Zurich, we analyse plasmid transmission efficacy using in vitro and mouse models. More information is available here.
- The Swiss Pathogen Surveillance Platform (www.spsp.ch): collaborating with the Universities and University Hospitals of Basel, Geneva and Lausanne, VetSuisse (University of Bern and Zurich) and the Swiss Institute for Bioinformatics, we are constructing an interoperable molecular and classical epidemiological database for WGS and metadata sharing for (multidrug resistant) pathogens. More information is available here.
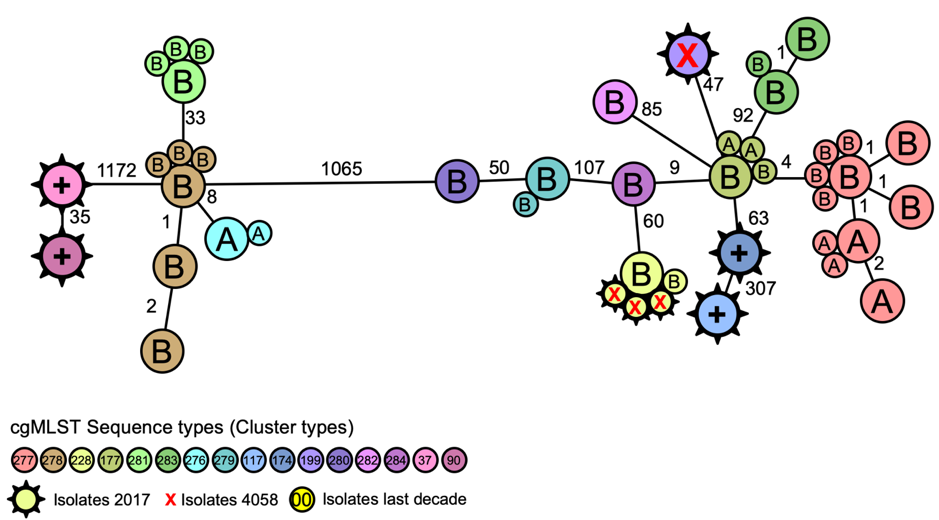
Figure 1. Core genome MLST (cgMLST) analysis of Legionella pneumophila in two air-conditioner cooling towers (A and B) and clinical isolates from patients (x) during an outbreak investigation. https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2019.24.4.1800192
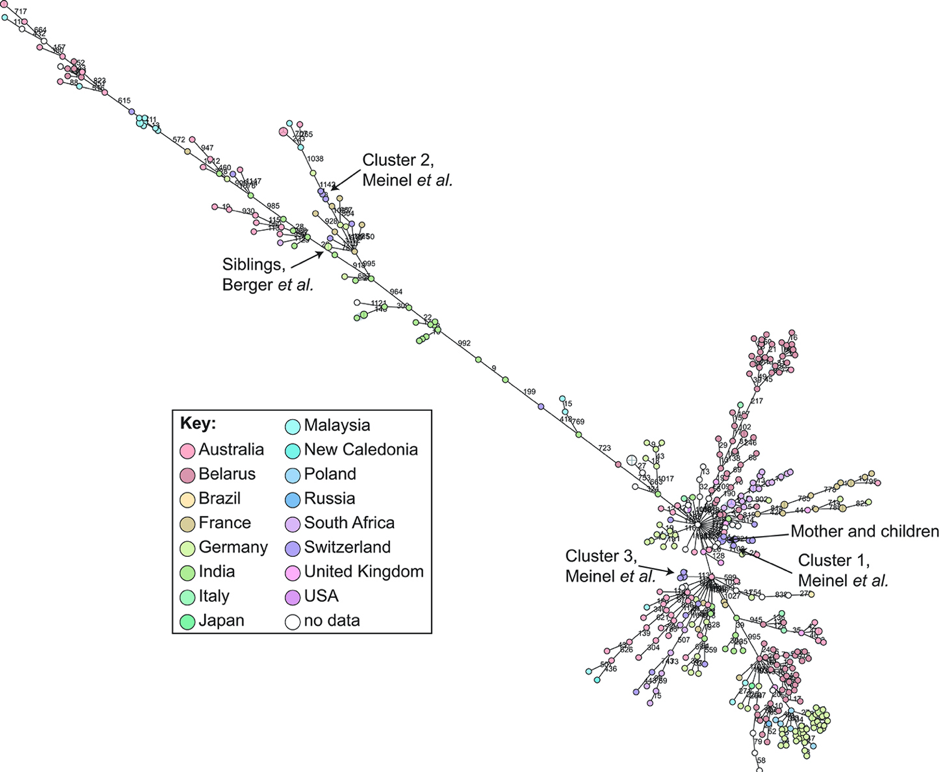
Figure 2. cgMLST analysis of Corynebacterium diphtheriae whole genome sequences from around the globe. https://doi.org/10.3389/fpubh.2019.00235
Viral transmission models
We use human influenza viruses and most recently SARS-CoV-2 to study transmission events in the context of local outbreaks and global transmission. For both viruses, we have established whole genome sequencing and analysis pipelines and humoral immune assays. Together with our collaborating partners, we explore transmission events and spatio-temporal dynamics across the Basel region, and investigate viral evolution in clinically relevant contexts, such as treatment of hospitalized patients.
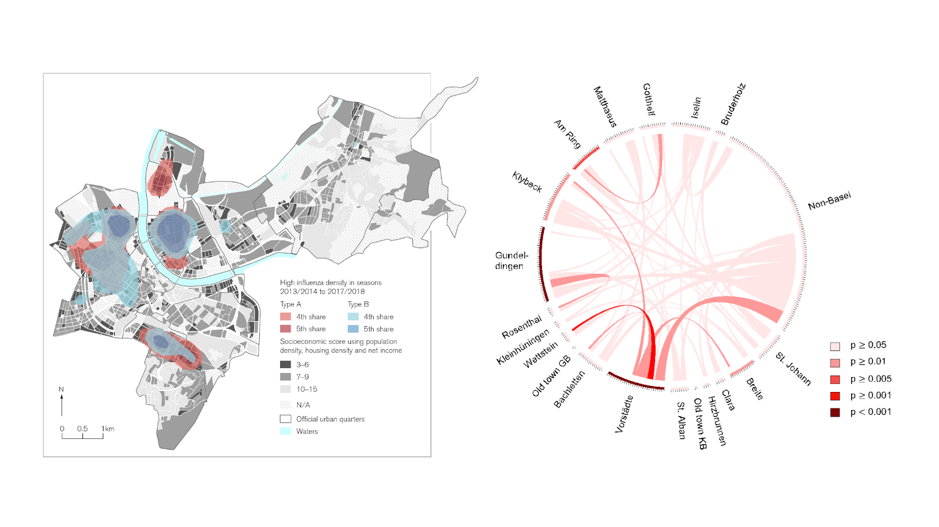
Figure 3. Left: PCR confirmed Influenza cases in Basel over the last five years visualized as a Kernel density plot (collaboration with Prof. Schneider-Sliwa, Humangeography, University of Basel). Right: Whole genome sequencing based data showing the exchange of highly similar influenza strains (red color intensity) between city quarters. https://www.biorxiv.org/content/10.1101/2020.04.03.023135v1
Retention and Displacement of ESBL E. coli
Antibiotic resistance is a severe threat to contemporary medicine. Therefore, we urgently need effective approaches to fight multidrug resistant pathogenic bacteria such as multidrug resistant E. coli. In 2017 the World Health Organization (WHO) classified the E. coli strains which express extended spectrum beta lactamases (ESBL E. coli), priority 1 due to their rise world-wide. So far there are no effective means to prevent the spread of ESBL E. coli.
Our project develops a radically new approach to fight ESBL E. coli. We hypothesize that ESBL E. coli can be efficiently displaced from the patient’s gut by applying well-defined microbials. Our approach roots in the observation that >50% of the colonized people spontaneously lose their colonizing ESBL E. coli strains within 6-18 months. We assume it to be due to the incidental ingestion of competitive, ideally pan-sensitive strains. Such strains are natural microbials and could provide a powerful means to stop the spread of ESBL E. coli.
- Firstly, we will perform a prospective observational single centre clinical cohort study to monitor ESBL E. coli colonization dynamics in the human gut. We enrolled 40 travelers to India and sampled their stool and blood at well-defined timepoints over one year. On these samples we are going to deploy shotgun metagenomics to obtain a broad and yet detailed picture of microbial dynamics over time and of the motility of genetic elements within the entire gut microbiota. This will give us an unprecedented picture of how microbe-microbe interactions shape the microbial successions in the gut, and on how the fading of ESBL strains over time takes place. Importantly, shotgun metagenomics will allow us to narrow down key displacer-pan-sensitive E.coli candidates for further testing. In parallel we will generate a systematic strain collection of ESBL E. coli isolates and their corresponding pan-sensitive E. coli strains. Each strain will undergo whole genome sequencing.
- In the second phase we will test our reconstructed colonisation dynamics, by testing the displacing ability of single candidate isolates in a mouse model. These pre-clinical experiments will establish whether pan-sensitive E. coli isolates from human participants can swiftly replace ESBL E. coli within the mammalian gut. Such natural strains would be the basis for future decolonization trials in ESBL E. coli positive humans.
- Finally our findings on ESBL displacement in humans and mice will be used to produce a colonization-displacement model that could serve to explain and possibly predict ESBL displacement, ultimately providing concrete indications for clinicians for an effective treatment.
Applications to routine diagnostics
We translate our findings to improve the speed, precision, and cost efficacy of routine diagnostic tools, and to reduce transmission of virulent and resistant pathogens.A key focus is on Matrix assisted laser desorption ionization - time of flight mass spectrometry (MALDI-TOF MS), which has revolutionized bacterial species identification in microbiological diagnostics. Despite the success of this technique, important challenges remain, such as detection directly from patient specimens and correct identification of closely related species.
Specific projects include:
- Developing protocols to use MALDI-TOF MS directly from human specimens such as positive blood cultures and urine samples. We have shown that rapid direct identification of positive blood cultures significantly reduces the time for correct antibiotic treatment and improved the patient outcome.
- Investigating the ability of MALDI TOF MS to distinguish between Klebsiella species. In a national and international consortium of routine diagnostic laboratories, we have elucidated species specific markers, compared whole genome sequences from our labs and databases, and predicted ribosomal protein masses which lie between 2,000 and 20,000 Daltons which can be detected by MALDI-TOF MS.
- Improving MALDI-TOF MS protocols, as the accuracy and resolution of MALDI-TOF is highly dependent on the spectra quality. We are currently conducting an international external quality assessment (EQA) including more than 30 laboratories around the globe to explore the factors associated with improved quality.
- Determining antibiotic resistance categories (susceptible or resistant) directly from MALDI-TOF MS spectral data with a machine learning based approach.
- Determining the phylogenetic characteristics of invasive and non-invasive E. coli isolates from patients using MALDI-TOF MS and whole genome sequencing data, aiming to enable earlier relevant clinical interventions.
Humoral immunity to pathogens
Flu vaccination significantly reduces the burden of influenza infection in a population. In certain risk groups, however, vaccination success is limited. In two large independent SNF-funded multi-center studies, we are measuring the pre- to post-vaccine humoral immune response towards influenza vaccination, and have established a protocol to determine haemagglutination inhibition assays (paper Kaufmann L et al.). The immune response is correlated to clinical factors such as degree of immunosuppression but also to genotypic variants in genes modulating the B-cell functionality. We also use computational modelling to understand the factors influencing the immune response.
Projects include:
- Cohort of solid organ transplant recipients vaccinated against influenza (STOP-FLU trial, SNF IICT grant)
- Cohort of hematopoietic transplant recipients vaccinated against influenza (SNF Ambizione grant)
Past Research Focus
Impact of Interferon lambda signaling on B-cell functions
In this SNF Ambizione funded project, we explored the impact of Interferon lambda on B-cell functions, especially in the context of influenza vaccination in immunosuppressed patients. We determined the specific gene expression patterns induced by Interferon lambda in synergy with IgM driven stimulation (PhD thesis, Mohameedyaseen Syedbasha). In a SystemsX funded iPhD project, we used mathematical modelling to understand the humoral immune response induced by influenza vaccination in more detail (PhD thesis, Janina Linnik).
Humoral immunity to pathogens
Flu vaccination significantly reduces the burden of influenza infection in a population. In certain risk groups, however, vaccination success is limited. In two large independent SNF-funded multi-center studies, we are measuring the pre- to post-vaccine humoral immune response towards influenza vaccination, and have established a protocol to determine haemagglutination inhibition assays (paper Kaufmann L et al.). The immune response is correlated to clinical factors such as degree of immunosuppression but also to genotypic variants in genes modulating the B-cell functionality. We also use computational modelling to understand the factors influencing the immune response.
Projects include:
- Cohort of solid organ transplant recipients vaccinated against influenza (STOP-FLU trial, SNF IICT grant)
- Cohort of hematopoietic transplant recipients vaccinated against influenza (SNF Ambizione grant)
Transmission of bacterial pathogens
In various projects, we have described:
- a Switzerland-wide nosocomial outbreak of Burkholderia stabilis
- an outbreak of Methicillin-resistant Staphylococcus aureus in refugees
- an outbreak of Vancomycin resistant Enterococcus in the region of Bern
- new species e.g. Mycobacterium basiliense
- Corynebacterium diphtheriae in refugees
- an outbreak of Legionella pneumophila showing links to air-conditioner cooling towers
- unusual cases including Blastobotrys raffinosifermentans
- Shigella sonnei as a sexually transmitted pathogen
- molecular epidemiological links of Campylobacter spp. isolates from human infections, chicken meat and environmental samples (in preparation, Seth-Smith HMB et al.)
- ongoing evolution of Chlamydia trachomatis LGV (in preparation, Seth-Smith HMB et al.)
Collaborations
We are proud of our past and ongoing collaborations locally, nationally and globally including researchers from the following institutes:
University Hospital Zürich
Biozentrum, University of Basel
D-BSSE, ETH Zurich
Department of Environmental Systems Science, ETH Zurich
Institute of Microbiology, ETH Zürich
Department of Health Sciences and Technology, ETH Zurich
University Hospital Basel
Swiss Tropical and Public Health Institute
Cantonal laboratory, City of Basel
Institute for Food Safety and Hygiene, University of Zurich
Swiss Pathogen Surveillance Platform
Funding
Research Grants:
- Swiss National Science Foundation
- Swiss Personalized Health Network (SPHN) & Personalized Health Related Technologies (PHRT)
- Gebert Rüf Foundation
- Rockefeller Foundation
- OAK Foundation
- Bangerter Rhyner Foundation
- Seerave Foundation
Supported by core funding from the University of Zurich